scatterPlot Plot GO terms as scattered points.
scatterPlot.RdscatterPlot Plot GO terms as scattered points.
scatterPlot(
simMatrix,
reducedTerms,
algorithm = c("pca", "umap"),
onlyParents = FALSE,
size = "score",
addLabel = TRUE,
labelSize = 3
)Arguments
- simMatrix
a (square) similarity matrix.
- reducedTerms
a data.frame with the reduced terms from reduceSimMatrix()
- algorithm
algorithm for dimensionality reduction. Either pca or umap.
- onlyParents
plot only parent terms. Point size is the number of aggregated terms under the parent.
- size
what to use as point size. Can be either GO term's "size" or "score".
- addLabel
add labels with the most representative term of the group.
- labelSize
text size in the label.
Value
ggplot2 object ready to be printed (or manipulated)
Details
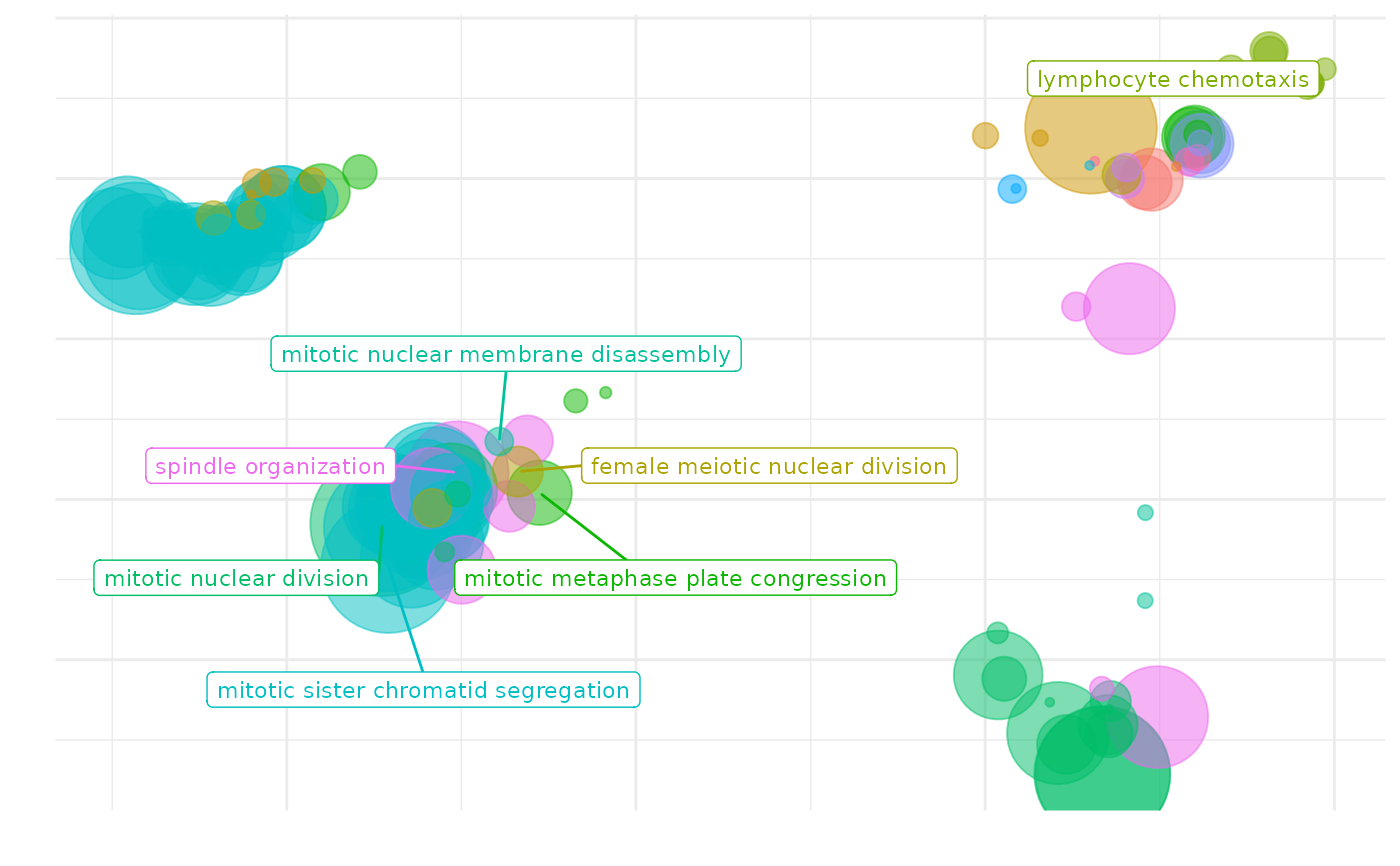
Distances between points represent the similarity between terms. Axes are the first 2 components of applying one of this dimensionality reduction algorithms: - a PCoA to the (di)similarity matrix. - a UMAP (Uniform Manifold Approximation and Projection,[1]) Size of the point represents the provided scores or, in its absence, the number of genes the GO term contains.
References
[1] Konopka T (2022). _umap: Uniform Manifold Approximation and Projection_. R package version 0.2.8.0, https://CRAN.R-project.org/package=umap.
Examples
go_analysis <- read.delim(system.file("extdata/example.txt", package="rrvgo"))
simMatrix <- calculateSimMatrix(go_analysis$ID, orgdb="org.Hs.eg.db", ont="BP", method="Rel")
#> preparing gene to GO mapping data...
#> preparing IC data...
scores <- setNames(-log10(go_analysis$qvalue), go_analysis$ID)
reducedTerms <- reduceSimMatrix(simMatrix, scores, threshold=0.7, orgdb="org.Hs.eg.db")
#> 'select()' returned 1:many mapping between keys and columns
scatterPlot(simMatrix, reducedTerms)
#> Warning: ggrepel: 10 unlabeled data points (too many overlaps). Consider increasing max.overlaps